Direct Sequencing Service
What is it?
This is a sequencing service we provide for scientists who have experience with sequencing and are doing regular sequencing with the platform without needing much support from us.
- We suggest that the request is send at least 5 days in advance of the pool being ready (ideally a week) so we can plan accordingly (kit, sequencing time slot, check files) and address your request asap when the pool is in the freezer
- Each submission is independant and typically corresponds to one sequencing run
- When you submit a request requiring a sequencing kit from the platform, you are buying this kit. If the submission is cancelled or postponed and the kit can’t be used, charges are on you. For frequently used kits that we keep in stock, it is generally not a problem to be used for another submission though.
- If delay in library prep, a sequencing request can be postponed to the following week, but not more. In that of a longer delay, the submission is cancelled and resubmitted when the libraries are close to ready.
How does it work?
- You fill the sequencing request form here
- We have tried to explain all the fields and details in the form directly
- After you submit the request, you will receive your submission by email (on the email you have filled in the form)
- This email includes your unique submission ID
- You can now create your folder and place the QC and samplesheet (as indicated in the email) - remember to follow the instructions here
- We validate the submission
- When we receive the request, we look at it and check if everything is clear (or ask questions if not)
- We schedule the sequencing with the first availability from the date you have mentioned that the samples are ready
- We check that we have the required files at least the day before the scheduled sequencing date
- You place the submission pool in the freezer
- As indicated in the email, you label the tube with submission ID, library concentration, name or initials and date of library submission to our freezer.
- min. 45 uL at 1nM
- min. 20ul at 4nM (fully QCed library pools only)
- min. 10ul at 10nM (pooled libraries for platform QC)
- As indicated in the email, you label the tube with submission ID, library concentration, name or initials and date of library submission to our freezer.
During that process, if there is any question or change, please email us at genomics@sund.ku.dk mentioning the submission ID.
If you have any suggestions to improve this process, please let us know.
How many base pairs / cycles
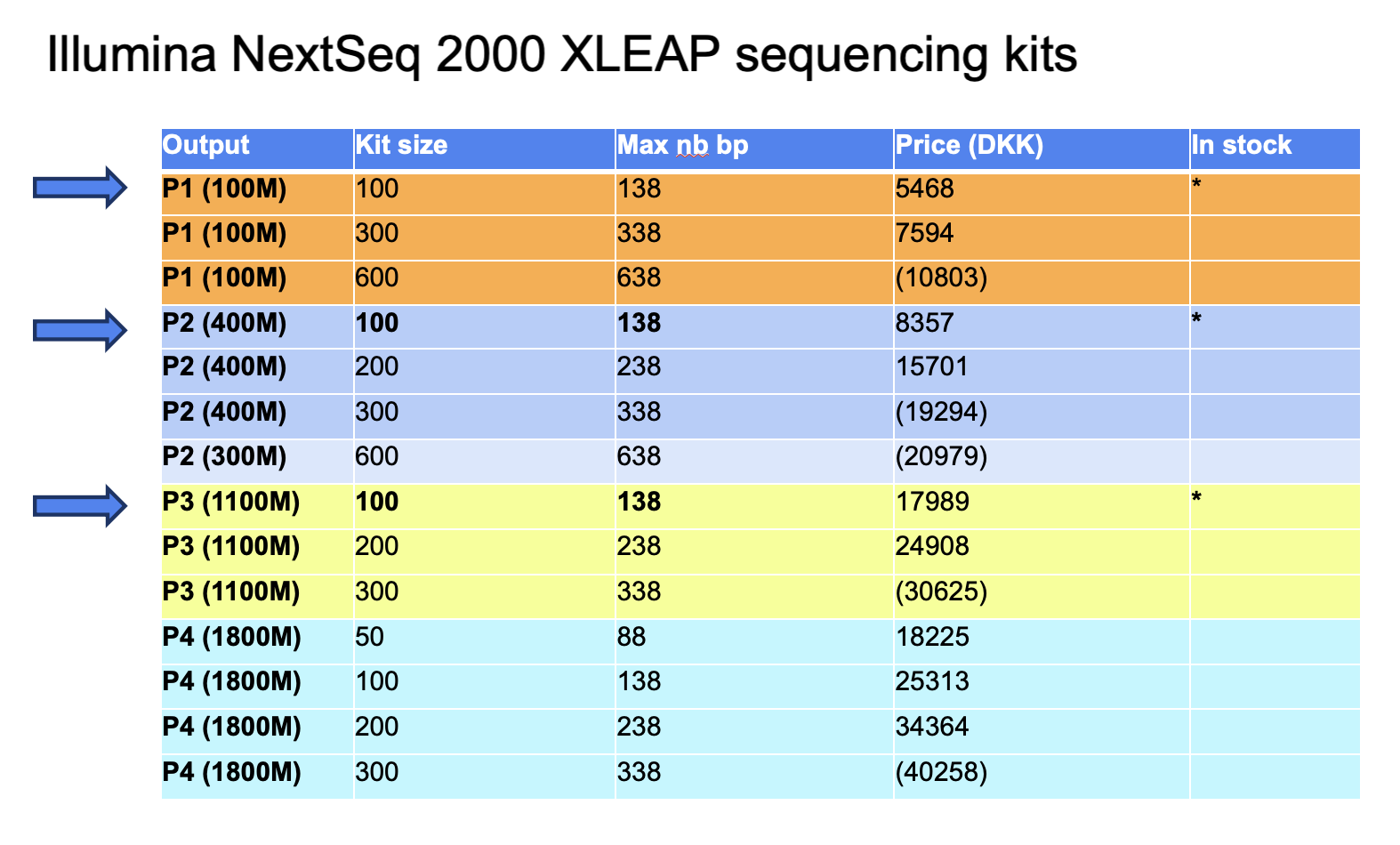
- The number of cycles availble for sequencing is determined by the kit and it reagent size. For Illumina NextSeq 2000, the sequencing kits and sizes are listed here (with their price).
- The kits in fact have more reagents than what is advertised on the product page and can run more cycles than the kit size. You can find the max number of cycles possible for each kit listed here.
- The max number of cycles can be distributed as you want between read 1, read 2, index 1, index 2
- For example
- a P2 v3 kit with 100 cycles (max=138) could be used
- 130 bp on read 1, 8 bp on index 1
- 61 on read 1, 61 on read 2, 8 bp on index 1, 8 bp on index 2
- 28 on read 1, 90 on read 2, 10 bp on index 1, 10 bp on index 2
- a P3 kit with 50 cycles (max=88) could be used
- 80 bp on read 1, 8 bp on index 1
- 41 on read 1, 41 on read 2, 6 bp on index 1
- a P2 v3 kit with 100 cycles (max=138) could be used
- Find a summary table here:
Go back to the Genomics Platform home